DNA Quantification with the UV Open Colorimeter
In this post we share some information on using the UV Open Colorimeter with a 255nm UV LED board for nucleic acid measurements.

Many widely-used molecular biology techniques require using accurate quantities of nucleic acids, for example qPCR/RT-PCR, cloning/bacterial transformations, in-vitro translation and sequencing. Instrumentation for quantifying nucleic acids is crucial for molecular biology and biochemistry research and educational labs.
The two most common methods for nucleic acid quantification are ultraviolet (UV) light absorbance (UV spectrophotometry) and fluorescence assays. UV spectrophotometry in particular is a very simple lab analytical method, requiring only access to a UV-Vis spectophotometer and no additional reagents or dyes. Substituting the spectrophotometer for an affordable, open hardware instrument like the UV Open Colorimeter makes this analytical method even more accessible.
We have previously shared methods for protein quantitation with the UV Open Colorimeter and a 275nm LED board. UV light absorbance by proteins is due to the Tyrosine, Pheynylalanine, and Tryptophan rings absorbing maximally at 280nm.
In this post we share a method for DNA quantification with the Open Colorimeter and a new 255nm LED board.
Nucleic acids have a specific UV absorbance peak due to the conjugated double bonds in the nucleotide bases (purine and pyrimidine rings) which have a specific absorption peak at 260nm. From the literature, we know the average molar extinction coefficient for double-stranded DNA at 260nm is 0.020 (μg/mL)−1 cm−1.
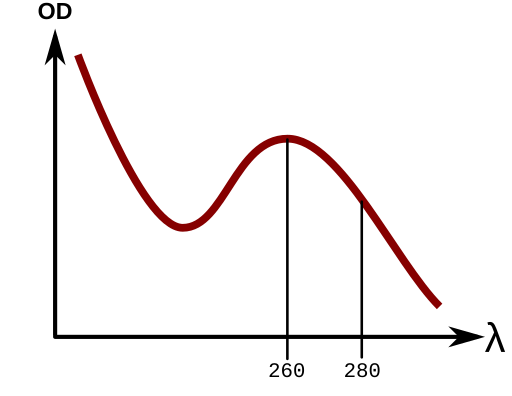
Using Beer's Law, you can accurately determine the concentration of nucleic acids from the molar extinction coefficient. Molar extinction coefficients and values for dsDNA, ssDNA and RNA when absorbance =1 are shown below:
- dsDNA, ε = 0.020 (μg/mL)−1 cm−1 | A260 of 1.0 = 50 μg/mL
- ssDNA, ε = 0.027 (μg/mL)−1 cm−1 | A260 of 1.0 = 33 μg/mL
- ssRNA, ε = 0.025 (μg/mL)−1 cm−1 | A260 of 1.0 = 40 μg/mL
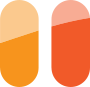
We recently designed a 255nm UV LED board for nucleic acid UV quantitation measurements with the UV Open Colorimeter. This board uses a 255nm LED on our current controlled LED board design. The board also has two 4-pin JST SH connectors for connecting to the UV light sensor and PyBadge.
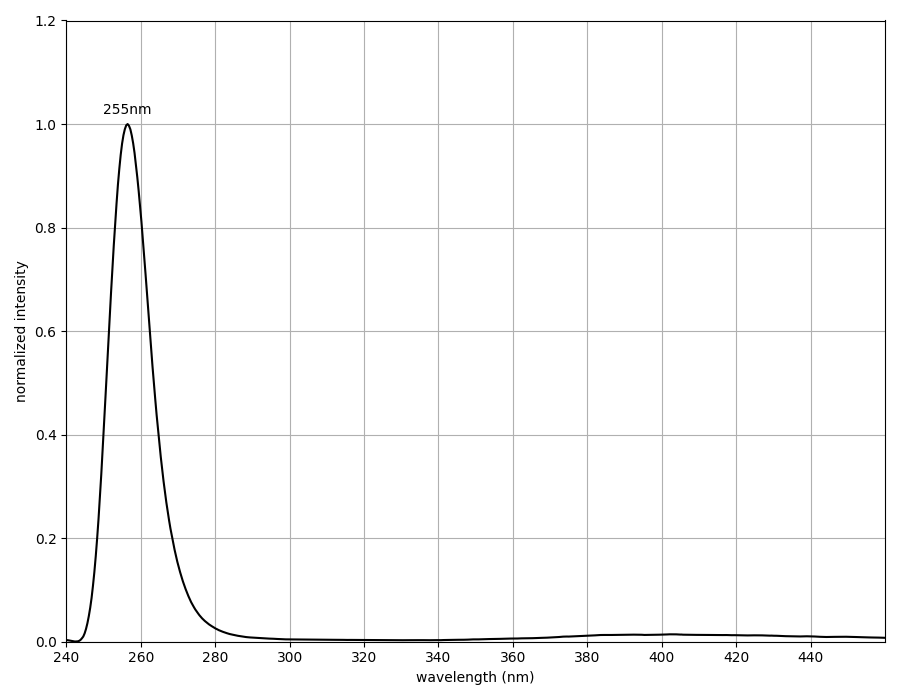
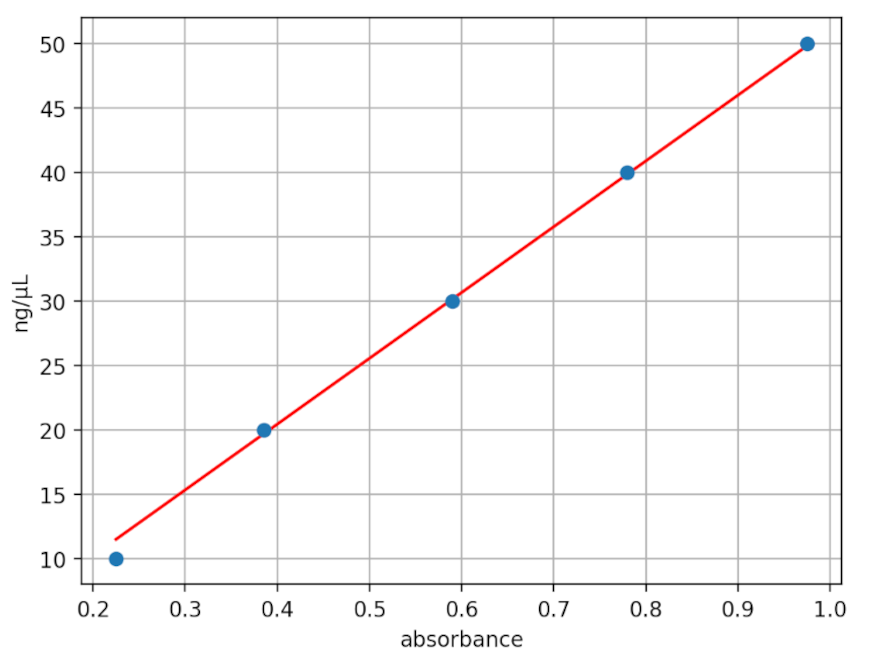
We characterized dsDNA absorbance with the Open Colorimeter at 255nm (A255) as described in the Supplemental Information at the end of this post. The graph below shows the measured absorbance data using the UV Open Colorimeter and dsDNA standard (blue data) compared to the expected values using the average extinction co-efficient for dsDNA (red data).
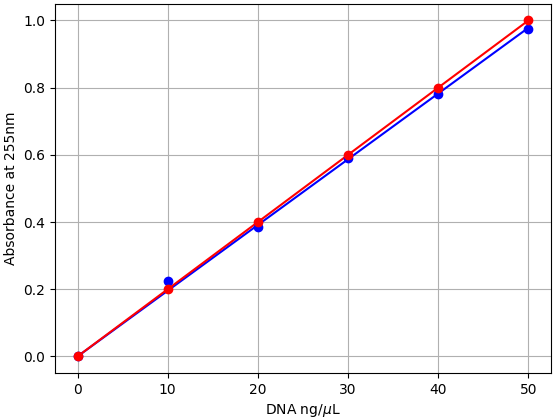
- Absorbance is linear over the 0-50 ng/µL dsDNA range tested
- dsDNA molar extinction coefficient at A255 with the UV Open Colorimeter is 0.0196 (μg/mL)−1 cm−1
- This is very close (2% difference) to the average dsDNA reported value
- Calibration data has now been added to the UV Open Colorimeter firmware and can be used to quantify a DNA sample as described in the method below
- There is more information about generating the calibration data for the Open Colorimeter in the Supplemental Information section at the end of this post
Methods: dsDNA Quant with UV Open Colorimeter
The UV Open Colorimeter now ships with a dsDNA quantification method as one of the default methods. Below are instructions for using this method to quantify dsDNA in a sample.
Materials
To use this method you will need the following materials:
- UV Open Colorimeter with 255 nm UV LED installed
- UV transparent micro cuvettes - 15 mm center height.
- 4 x UV transparent cuvettes are included with the UV Open Colorimeter. More can be purchased from online stores
- DNA sample for quantitation
- Distilled water or buffer for diluting DNA sample if necessary
- 0.2-1.0 mL adjustable micropipettes

Procedure
General instrument navigation instructions (main menu button, blanking and gain & integration settings) are in the Product Guide.
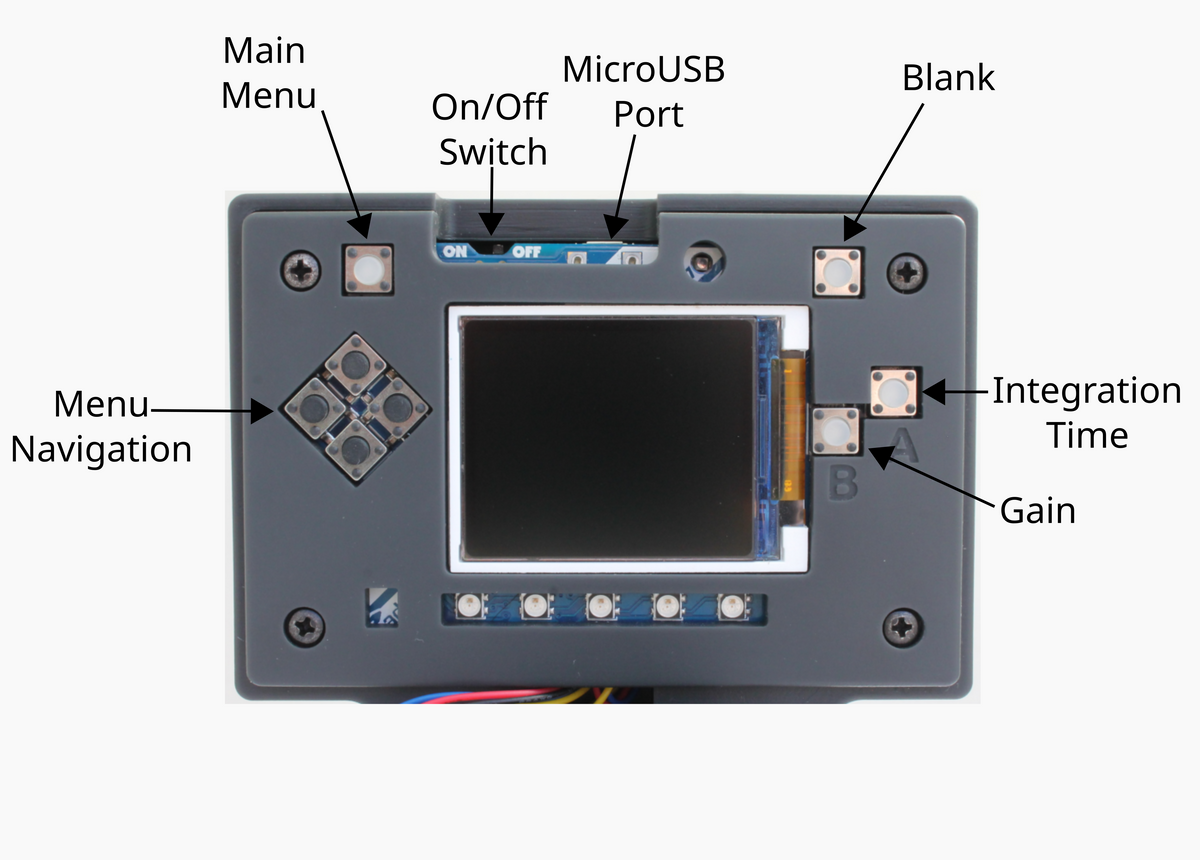
- With the 255nm UV LED installed, switch on the UV Open Colorimeter. After the splash screen, the first screen is the default Absorbance screen as shown below

- Go to the Main Menu (top left button) and navigate ⇩ to the dsDNA method.


- Blank the cuvette with a zero DNA sample (e.g. distilled water of buffer) using the top right "blanking" button

- The colorimeter is now ready for measuring your sample. Place your cuvette with DNA sample into the cuvette holder. DNA concentration will be displayed in ng/μL.

Supplementary Information
Calibrating the Open Colorimeter using the oc-calibration app
In this supplementary section we provide some additional insight as to how we prepared the dsDNA quantification method that is pre-installed on the instrument. This information can also be used if you want to prepare a new calibration with a nucleic acid standard. Currently we have not tested other nucleic acids, for example ssDNA or RNA.
UV Light Sensor Settings: The UVC measurement channel was used. Gain and integration time were typically set at 128 x gain and 256 ms respectively.
We prepared a 6-point dsDNA dilution series using a 100ng/uL dsDNA sample. We have been using a dsDNA 100ng/uL Biotium AccuGreen™ Standard 2 ($48 for 1mL). All samples were measured in a UV-transparent cuvette. After blanking with distilled water, we measured absorbance values at 255nm. We repeated measurements for two different batches of DNA standards purchased from Biotium in July and August 2024, shown as Standard 1 and Standard 2 respectively.
| DNA Conc. |
DNA Standard | Distilled H₂O | Standard 1 A255 | Standard 2 A255 | Avg. A255 |
|---|---|---|---|---|---|
| 0.0 ng/µL | 0 µL | 300 µL | 0.00 | 0.00 | 0.00 |
| 10 ng/µL | 30 µL | 270 µL | 0.23 | 0.22 | 0.225 |
| 20 ng/µL | 60 µL | 240 µL | 0.38 | 0.39 | 0.385 |
| 30 ng/µL | 90 µL | 210 µL | 0.590 | 0.590 | 0.590 |
| 40 ng/µL | 120 µL | 180 µL | 0770 | 0790 | 0780 |
| 50 ng/µL | 150 µL | 150 µL | 0.97 | 0.98 | 0.975 |
Next we used the oc-calibration app to generate the json file from the averaged absorbance values. Instructions for using the oc-calibration are in our Product Guide here: https://blog.iorodeo.com/oc-calibration-app/.
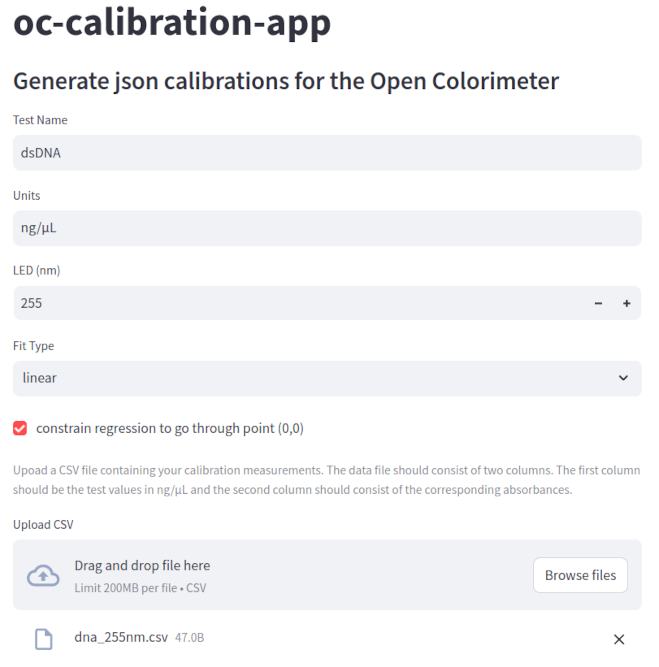

Screenshots from using the oc-calibration-app to generate dsDNA json calibration data
Finally, the json calibration data was downloaded onto installed into the calibration.json on the UV Open Colorimeter firmware. This generates the method on device which is listed as "dsDNA" in the main menu.
UV Open Colorimeter - Store Link
You can learn more about our UV LEDs and UV Sensor in the Open Colorimeter Product Guide.
Bookmarks





Comments ()